About This File
Whole genome sequence, SNP chips and pedigree structure: Building demographic profiles in domestic dog breeds to optimize genetic trait mapping
Authors: Dayna L. Dreger, Maud Rimbault1, Brian W. Davis1, Adrienne Bhatnagar1, Heidi G. Parker, Elaine A. Ostrander
Key Words: population, homozygosity, canine, inbreeding
SUMMARY STATEMENT
"Successful application of whole genome sequencing and genome-wide association studies for identifying both loci and mutations in canines is influenced by breed structure and demography, motivating us to generate breed-specific strategies for canine genetic studies."
ABSTRACT
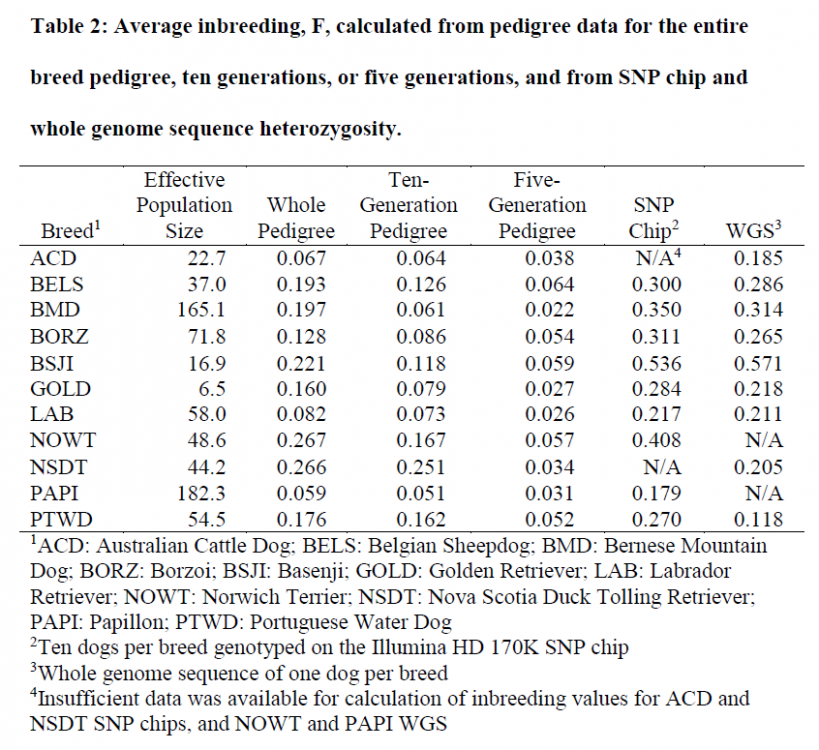
"In the decade following publication of the draft genome sequence of the domestic dog, extraordinary advances with application to several fields have been credited to the canine genetic system. Taking advantage of closed breeding populations and the subsequent selection for aesthetic and behavioral characteristics, researchers have leveraged the dog as an effective natural model for the study of complex traits, such as disease susceptibility, behavior, and morphology, generating unique contributions to human health and biology. When designing genetic studies using purebred dogs, it is essential to consider the unique demography of each population, including estimation of effective population size and timing of population bottlenecks. The analytical design approach for genome-wide association studies (GWAS) and analysis of whole genome sequence (WGS) experiments are inextricable from demographic data.
We have performed a comprehensive study of genomic homozygosity, using high-depth WGS data for 90 individuals, and Illumina HD SNP data from 800 individuals representing 80 breeds. These data were coupled with extensive pedigree data analyses for 11 breeds that, together, allowed us to compute breed structure, demography, and molecular measures of genome diversity. Our comparative analyses characterize the extent, formation, and implication of breed-specific diversity as it relates to population structure. These data demonstrate the relationship between breed-specific genome dynamics and population architecture, and provide important considerations influencing the technological and cohort design of association and other genomic studies."
excerpt...
"The work presented here examines variables of inbreeding and homozygosity in a large and comprehensive set of dog breeds through parallel use of pedigree data, genome-wide SNP genotyping, and WGS. Specifically we compare data from extended pedigree analysis, genotyping with a SNP chip of 173,622 potential data points, and WGS with an average depth of 27.79X.
We found that each dog breed has a unique profile of genome diversity, varying by amount of total homozygosity as well as number and size of homozygous regions. Likewise, while we observe variation between members of the same breed, multiple individuals from a single breed can be combined to obtain an accurate reflection of breed-specific homozygosity and knowledge regarding fluidity of variation within breed confines. This allows us to define metrics that inform the design of canine genetic studies while also allowing us to develop an understanding of the intricate complexity of the diversity of dog breeds. Individual diversity metrics are provided for over 100 breeds as a resource for investigators in the field."
 Donate
Donate