"The Downside of Inbreeding"
The Downside of Inbreeding - It’s Time For a New Approach
by C.A. Sharp
First published in Double Helix Network News, Winter 1999
I am pulling together a presentation for the Embrark Canine Health Summit and came across an article I have cited before... an oldie but a goldie! This article is by the very knowledgeable C.A. Sharp, expert and person behind ASGHI (Australian Shepherd Genetics and Health Institute) and one of our collaborating partners.
Good news - she gives a great coverage of this important topic. Of concern - this was written in 1999 and it is still a hot topic today. READ it here: https://www.ashgi.org/home-page/genetics-info/breeding/breeding-genetic-diversity/the-downside-of-inbreeding
Embark has been regularly publishing Coeffiecients of Inbreeding (COI's) on genetic samples from various breeds on its Embark for Breeders facebook page.
These are genomic NOT pedigree based COIs (learn about the difference here) and understandably but unfortunately we do not know how many dogs were included in the calculations. Presumably it is an international sample. Notwithstanding this limitation, these are useful to consider.
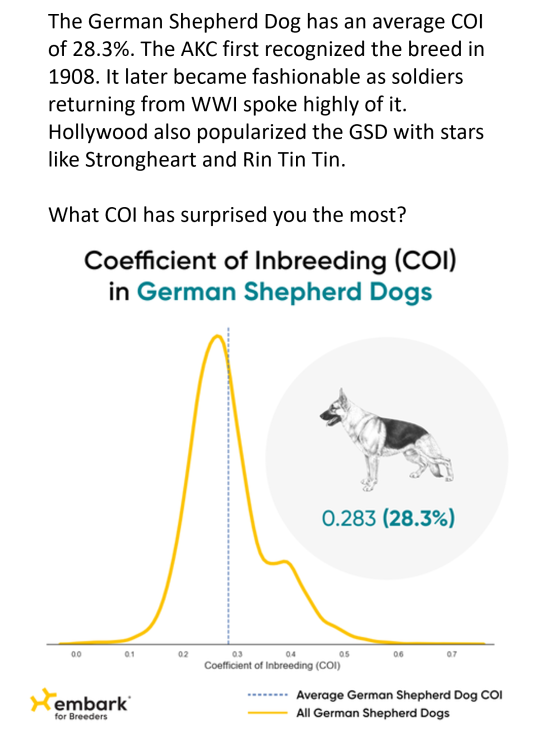
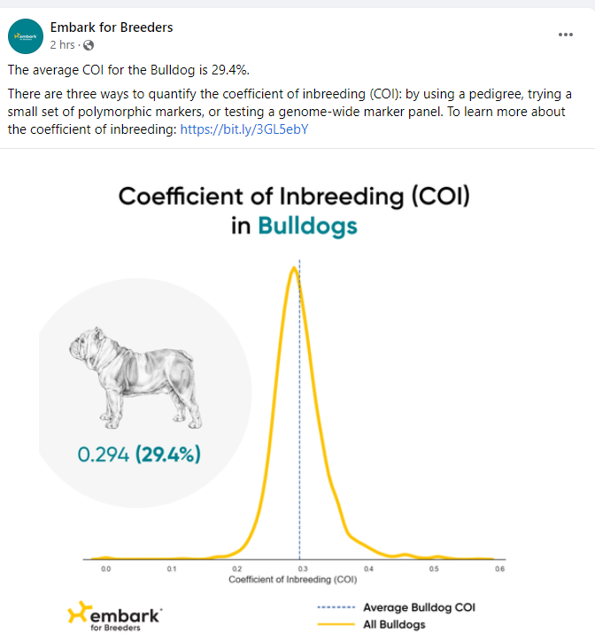
Firstly, remember that a brother-sister mating results in a COI of 25%. That is inbreeding. Look at these values from Embark, as examples.
Both the German Shepherd Dog and the Bulldog have average COIs above that level... meaning that many of the dogs have values that are higher still. On average, dogs with a COI >25% share more genetic material from common ancestors than would arise from a brother-sister mating.
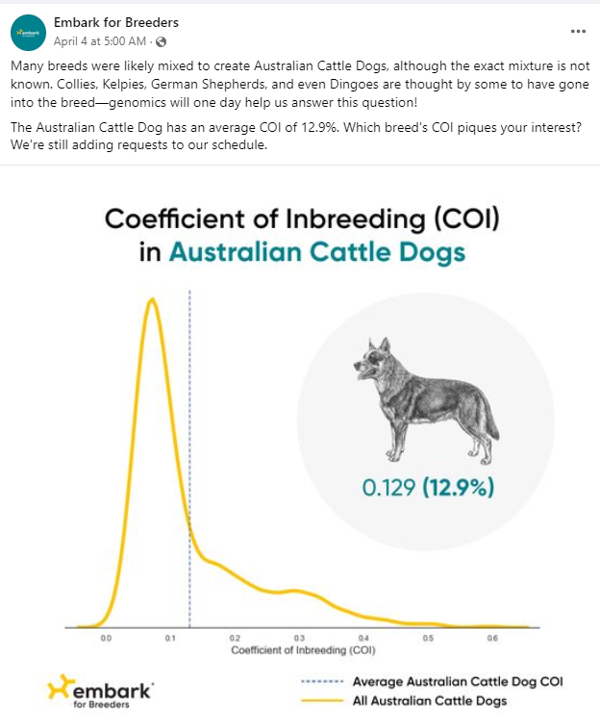
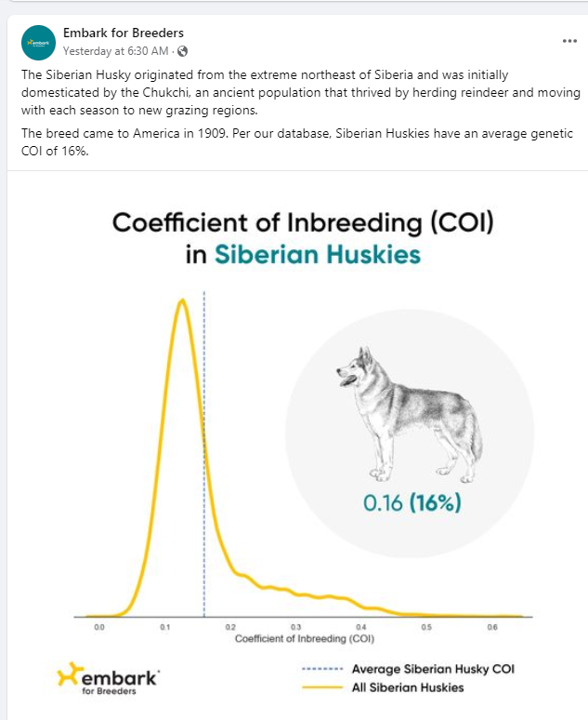
Results for the Cattle Dog and the Husky below show lower values for COIs. Not surprisingly, perhaps, given a focus on performance in these breeds?
Protecting the future of your breed depends on Genetic Diversity. Make sure you understand how breeding practices like line-breeding result in reduced diversity and over time may create health problems like poor reproductive capacity, lowered longevity and more.
Other reading: Linebreeding vs. Inbreeding – Let’s be perfectly clear.
 Donate
Donate
2 Comments
Recommended Comments